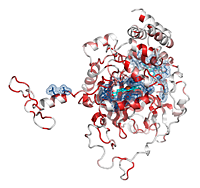
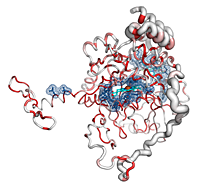
Catalases are tetrameric haeminic enzymes that decompose hydrogen peroxide into water and oxygen. They serve to protect cells from the toxic effects of hydrogen peroxide.
We will use ENDscript to decipher features in the crystal structure of Proteus mirabilis catalase (PDB entry 2CAH).
The two fundamental residues of the haem-iron catalytic site are a distal histidine and a proximal tyrosine. A unique methionine sulfone is observed in the distal site of Proteus mirabilis catalase. Another peculiarity of this bacterial catalase is its ability to bind nicotinamide-adenine-dinucleotide phosphate (NADPH) to prevent inactivation by hydrogen peroxide.
The user clicks the  icon of the ENDscript form, then types icon of the ENDscript form, then types 2CAH and presses the Retrieve data buttonThe query 2CAH is pre-analyzed by ENDscript
- The following tooltip appears under the panel Keeping contacting hetero-compounds :
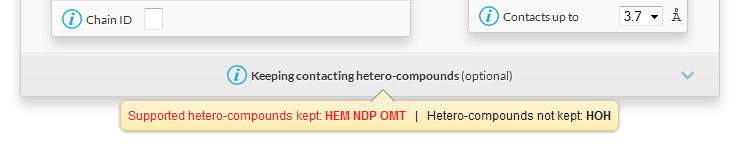
HEM is the bound haem, it will be considered as a ligand in ENDscript.NDP is the bound NADPH, it will be considered as a ligand in ENDscript.OMT is the methionine sulphone residue of the polypeptide chain, it will be considered as a protein residue.HOH are water molecules, they will be removed in ENDscript unless the name HOH is written in the tabular form Keeping contacting hetero-compounds (click on the panel to unroll it).
The user decides to keep all the default settings and presses the SUBMIT button
- The
2CAH sequence is extracted using SPDB.
- A BLAST search is performed against the PDBAA sequence database (derived from the PDB) with an E-value threshold of 1e-6.
- Sequences of homologous proteins found by the BLAST search are aligned using Clustal Omega.
- Secondary structure elements are extracted using DSSP and accessibility is calculated.
- Cα chains of homologous proteins are superimposed on that of 2CAH using ProFit.
- ESPript and PyMOL are used to render 2D and 3D information.
The ENDscript "RESULTS" pop-up window appears
- The user can click on the
2CAH hyperlink to be taken to its PDB summary page.
- The user can open the ESPript - Phase 1 PDF file ( download it [20 Kb] ) generated by ESPript and note that:
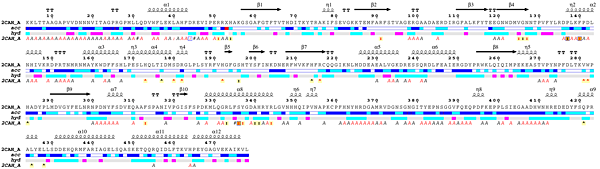
2CAH crystallizes with one monomer in the asymmetric unit, e.g. chain A (labeled 2CAH_A on the figure).
- The catalase structure has an α + β topology.
- Residues 1 to 3 are not observed and the
2CAH sequence starts at Lys4.
- The N-terminal region is involved in extensive protein:protein crystallographic contacts as indicated by italic
A letters below the sequence block. A red letter identifies a contact < 3.2 Å while a black letter identifies a contact between 3.2 and 5 Å.
- Leu31 is positioned along a 2-fold crystallographic axis as indicated by an italic
#.
- Leu40 is in contact with the haem group of a symmetric monomer as indicated by an italic colon
:.
- Asp44 is also in contact with the haem group of a symmetric monomer as indicated by an italic colon
: It is also involved in a protein:protein crystallographic contact as indicated by a blue frame.
- Arg51 binds the haem group of the monomer as indicated by a normal colon
: on a light yellow background.
- The non-standard residue at position 53 (labeled
X) is the distal site methionine sulfone.
- Phe140 may be a critical residue: it has close contacts with the haem group as indicated by the red colon
: on a yellow background. It is also involved in protein:protein contacts as indicated by the blue frame. Finally, it is involved in both crystallographic and non-crystallographic contacts as indicated by the orange background. The detailed list of contacts is accessible via the CNS hyperlink in the Tracing files tab of the "RESULTS" pop-up window. Using this CNS file, you can see that Phe140 is involved in both a crystallographic contact with Phe43 and in a non-crystallographic contact with the haem group.
- His173 binds tightly NADPH as indicated by a red caret
^.
- The user can open the ESPript - Phase 2 PDF file ( download it [61 Kb] ) generated by ESPript and note that:
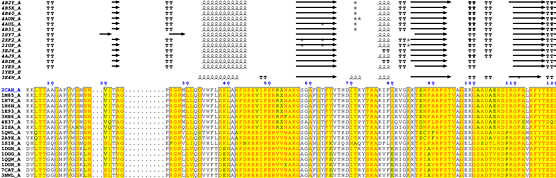
Excerpt from the ESPript phase 2 figure
- Secondary structure elements are well conserved in catalases.
- The essential distal His54 is substituted in some sequences to obtain an inactive protein.
- The user can open the PyMOL Sausage representation ( download it [7.2 Mb] ) to interact with the 3D structure.
- He can press on the CONTACTING_RES and SITES buttons in the right PyMOL control panel to observe the haem and the NADPH binding sites.
- He can observe that the Cα trace of the C-terminal region varies between catalases (the tube radius is proportional to the mean r.m.s. deviation between
2CAH and homologous catalases). Consistently, the C-terminal region is poorly conserved in sequence and colored in white (color ramp from white, low conservation, to red, identity).
- In contrast, the N-terminal region, which protrudes from the core of the protein, is surprisingly well conserved. In fact, this region is deeply buried in the biological tetramer. The user can click on the four BIOUNIT1 buttons in the right PyMOL control panel to generate the tetramer. The user can also see that the four haem groups are well buried in the tetrameric protein.
- The user can cross-check that the N-terminal Leu40 and Asp44 are in contact with the haem of a symmetry-related monomer.
- Finally, the user can click on the SOLV_SURFACE button in the right PyMOL control panel to observe the small channel that drives the hydrogen peroxide substrate from the protein surface to the haem distal site.
|