

|
|
November 18, 2025: Release of ENDscript v2.2 integrating the AlphaFold Protein Structure Database (with around one million entries from the ‘Model Organism Proteomes’, ‘Global Health Proteomes’ and ‘Swiss-Prot’ subsets). October 25, 2025: We are pleased to announce the release of our new web server: NESPript  It has been designed for the representation of multiple alignments of nucleic acid sequences, including the visualization of their secondary structures. |
| ENDscript Outputs |
|---|
|
Here are some examples of ENDscript output from selected PDB entries. Click on the thumbnails below to view the images in full size, or download the corresponding flat figures (in PDF format) and PyMOL session files for interactive 3D representations (PyMOL must be installed on your computer). |
| • Example #1 | |||
|---|---|---|---|
| PDB entry 3B4N : Crystal structure of pectate lyase PelI from Erwinia chrysanthemi | |||
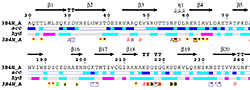
|

|
ESPript flat figure, phase 1 (PDF) | [12 Kb] |
| ESPript flat figure, phase 2 (PDF) | [13 Kb] | ||
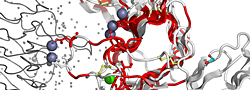
|
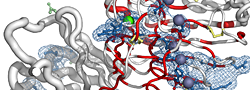
|
PyMOL 'Cartoon' session file | [3.2 Mb] |
| PyMOL 'Sausage' session file | [3.2 Mb] | ||
| • Example #2 | |||
|---|---|---|---|
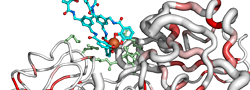
| PDB entry 2CHU : Crystal structure of a ferric enterobactin binding protein with a [{Fe(mecam)}2]6-bridge | |||

|

|
ESPript flat figure, phase 1 (PDF) | [12 Kb] |
| ESPript flat figure, phase 2 (PDF) | [18 Kb] | ||
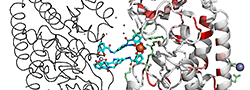
|
|
PyMOL 'Cartoon' session file | [2.8 Mb] |
| PyMOL 'Sausage' session file | [2.8 Mb] | ||
| • Example #3 | |||
|---|---|---|---|
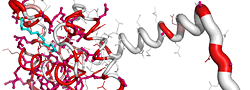
| PDB entry 1UPH : NMR structure of HIV-1 myristoylated matrix protein | |||
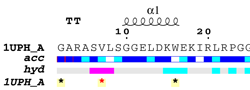
|
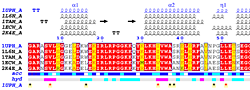
|
ESPript flat figure, phase 1 (PDF) | [8 Kb] |
| ESPript flat figure, phase 2 (PDF) | [12 Kb] | ||
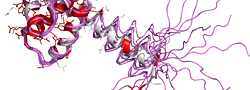
|
|
PyMOL 'Cartoon' session file | [2.6 Mb] |
| PyMOL 'Sausage' session file | [2.6 Mb] | ||
| • Example #4 | |||
|---|---|---|---|
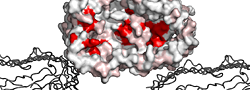
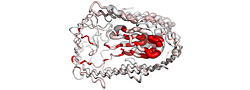
| PDB entry 3OHN : Crystal structure of the FimD translocation domain | |||

|

|
ESPript flat figure, phase 1 (PDF) | [14 Kb] |
| ESPript flat figure, phase 2 (PDF) | [16 Kb] | ||
|
|
PyMOL 'Cartoon' session file | [8.3 Mb] |
| PyMOL 'Sausage' session file | [8.3 Mb] | ||
|
|
© 2005-2026 The ENDscript authors & CNRS - Contact: espript@ibcp.fr ESPript is an SBGrid supported application |